A Chemist’s Toolkit for Biology
- D-CHAB
- LOC
- Highlights
Sometimes we must create something artificial to understand nature. Kathrin Lang, new professor of Chemical Biology at the LOC, develops special tools to do so, opening molecular doors that not only allow insights into existing complex biological systems, but also show what is possible beyond them.

„Du bisch a von Südtirol?“ (You are from South Tyrol too?) The ice is broken even before we are taking a seat between the five plants in this particular office. Two South Tyrolean women are sitting there at the table: one is interviewing, the other, who is the protagonist of this text, comes from Gries/Bolzano. Her name is Kathrin Lang and she is proudly showing with both hands how much the large cactus behind her has grown since it was given to her as a farewell gift at the TU Munich. In the summer of 2021, Kathrin Lang joined ETH Zurich as a full professor, with doctoral students, postdocs and several molecular tools in her luggage. Now at ETH, she wants to expand this chemical repertoire to take a closer look at the lives and action of the main players on the biological field: Proteins.
„Actually, I always wanted to study mathematics and solve scientific problems with it," Kathrin Lang recalls. However, she dropped out of her studies in technomathematics in Munich when she realized that optimizing ignition processes at BMW was definitely not her career plan and interest. She opted for chemistry in Innsbruck, where she eventually found her place in chemical biology. "In my doctorate, I carried out modified RNA synthesis for the first time and worked with ribosomes. These are the complex machines in the cell that translate our genetic information into proteins. Absolutely fascinating," Lang says.
Her enthusiasm led her to a postdoc position in Cambridge in 2009, with Venki Ramakrishnan, who received the Nobel Prize for his contributions in ribosome crystallography in the very same year. "His high-resolution structural images provided insights into the protein factories of the cell for the first time. That was exciting," Kathrin Lang recalls. But she missed the practical chemistry. So, she joined Jason Chin's group in Cambridge, where she first got in contact with synthetic biology, and eventually moved to TU Munich for a professorship in 2014. Since then, she and her team have been conducting research in the field of chemical and synthetic biology.

"What I cannot create, I cannot understand"
Richard Feynman once said: "What I cannot create, I cannot understand”. This principle is also followed in chemical and synthetic biology. It is an interdisciplinary subject at the interface of organic chemistry, biotechnology, molecular biology and engineering. „That makes it so fascinating,“ says Kathrin Lang, who is a huge fan of out-of-the-box approaches. "In my group, we develop chemical tools to create biological systems that don't occur like this in nature. We mimic them and modify them. This way, we can answer biological questions like: why does nature something in a certain way and not in another. At the same time, we can create things that are not possible in nature."
The field of application is broad. For example, bacteria can be modified so that they can break down plastics, enzymes can be generated to carry out new chemical reactions or antibodies can be produced that attack certain cancer cells. Kathrin Lang gets inspiration for her tools from exchanges with biological institutes or at biological symposia. She still thinks chemically, in terms of atoms, molecules and mechanisms, even when it comes to biological processes. Three biological questions are currently of particular interest to the Lang group and form now the pillars of their research at ETH Zurich: how can proteins be modified with biophysical probes, e.g. to make them visible? When and how do proteins interact with other proteins in the cell? And why and how are proteins modified after their synthesis, e.g. ubiquitinated?
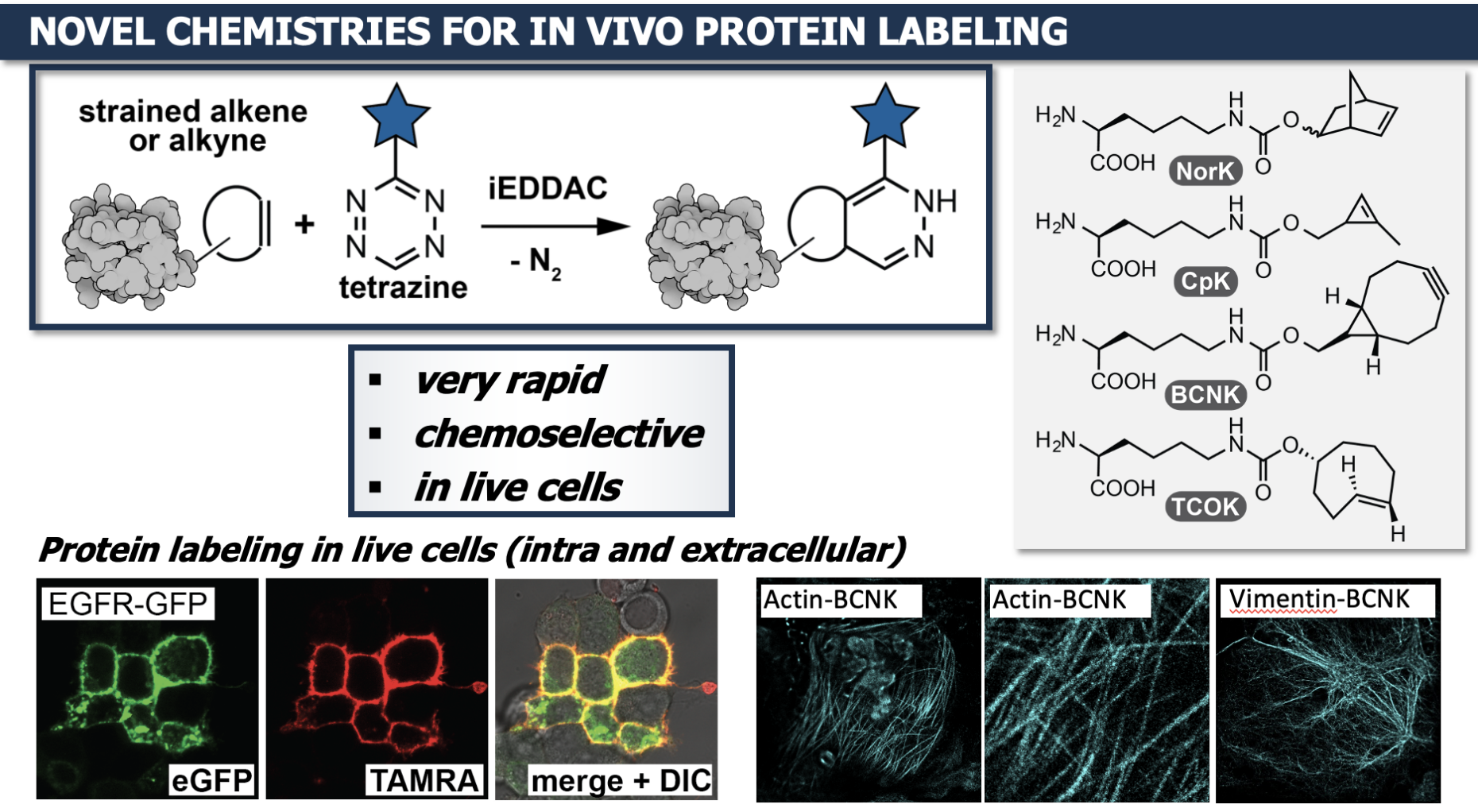
Beyond the 20 natural amino acids
"Every living being, from the simplest single-celled organism to us humans, uses the same 20 amino acids. They are the building blocks for our proteins and the order of these 20 building blocks is fixed in the genetic code," explains Kathrin Lang. "My group is working on extending the highly conserved genetic code to incorporate artificial amino acids beyond these natural 20 amino acids into proteins in living cells. This enables to endow proteins with new functional groups and with new physicochemical properties."
Kathrin Lang's lab is particularly interested in labeling, which means in this case: making proteins visible. Common fluorescent proteins such as GFP are often large and thus can influence the target protein. Instead, modified amino acids can be used. "These are functionalized with a kind of handle. In the living cell, the handle is supposed to react with its 'counterpart', which carries a fluorescent dye, a fluorophore. This way, you get a fluorescent protein," Lang explains. "You could use it, for example, in carcinogenic cells, to track what happens to a certain protein. Is it perhaps being transported to another compartment of the cell?" The development of such reactions is very complex, because the reaction between handle and fluorophore must be extraordinarily selective and it takes place in the most complex environment imaginable: in the living cell. "We can't just heat things up or apply toxic agents if a reaction doesn't work. Our tools have to work independently and in parallel to all that chemistry that is going on in living cells, and that's a lot." In the future, the group would like to introduce several different fluorophores. This could be used to study how proteins fold in the cellular milieu, but this is work in progress.
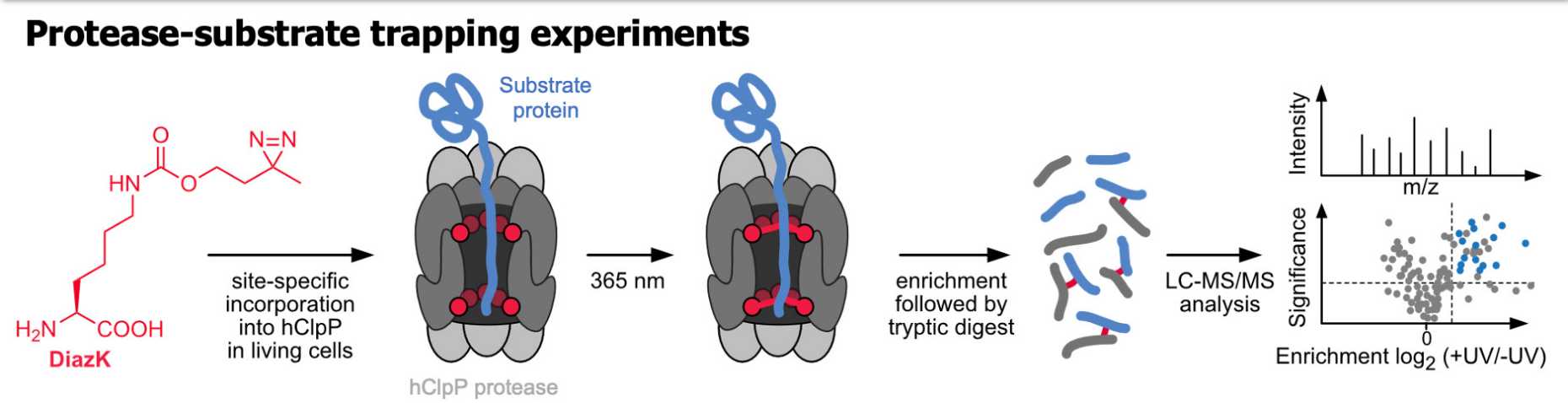
It is also important to understand that proteins do not act in isolation but interact with a variety of other proteins and biomolecules in their natural environment. To study this, Kathrin Lang's group has developed modified amino acids that can be incorporated into a protein and, when exposed to light, form a covalent bond with a protein nearby. "In mitochondria, for example, there is a protein degradation system that is known to be important in certain diseases; however, it was not clear which proteins are degraded in this system," Lang says. "So, we positioned our amino acids in this system. They allowed us to trap the proteins that were passing by and eventually identify them."
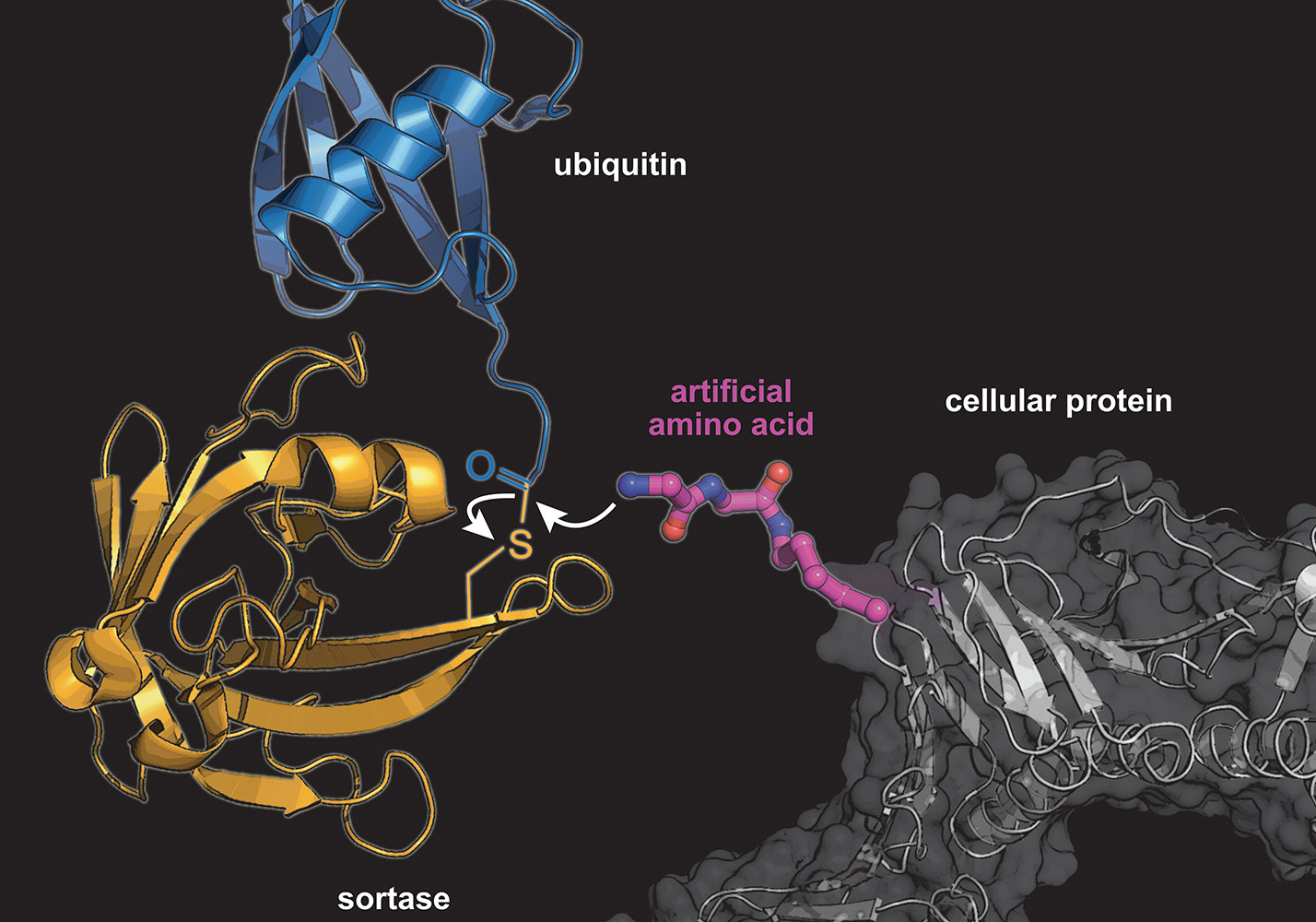
Furthermore, a particularly important biological regulatory mechanism is the modification of proteins after their biosynthesis. This includes, for example, ubiquitination, in which one or more small ubiquitin proteins are attached to a target protein. The type, position and number of ubiquitin molecules bound to a protein determine the stability, function and location of the protein within the cell. "Almost every cellular process is influenced by ubiquitination. Thus, malfunctions in the ubiquitination mechanism lead to many serious diseases such as cancer," explains Kathrin Lang. However, due to the complexity of ubiquitination, it is difficult to look inside such systems. "Therefore, we are developing methods to recreate these complex systems – both in the test tube and in the living cell. This will then allow us to study the effects of ubiquitination patterns, for example, on the development of cancer or neurogenerative diseases at the molecular level."
Broadly applicable tools
"The value of a tool is determined by its applicability," Lang emphasizes. "Our goal is that cell biologists, neurobiologists or biochemists will be able to reproduce and use our tools easily, and that our tools are able to shed light on biological processes that cannot be studied with more traditional approaches. We have already patented a few things. Our aim is to ensure that many of our tools can eventually be used as a kit with simple instructions for use in any biological lab."
The next step for Kathrin Lang will be to further expand her lab group – ideally in a way that everyone gets along well and the chemistry is right, she says, because the work here is very interdisciplinary and new thematic challenges at the border between chemistry and biology are always welcome. „In addition, after Corona and the stress of moving, I definitely have to climb some mountains in Switzerland and expand my circle of acquaintances here,“ Kathrin Lang adds with a laugh. The interview with the South Tyrolean, who is now packing away her recording device at the other end of the table, was already a good start for this.

Inaugural Lecture - Kathrin Lang
An expanded genetic code – adding new chemistries to biology
1 June 2022
Time: 17.15 – 18.30
Location: Zürich Zentrum HG F 30
Livestream: https://video.ethz.ch/live/events/live-1.html
Group website
Sources and further information
Kathrin Lang, Lloyd Davis, Stephen Wallace, Mohan Mahesh, Daniel J. Cox, Melissa L. Blackman, Joseph M. Fox, and Jason W. Chin (2012): Genetic Encoding of Bicyclononynes and trans-Cyclooctenes for Site-Specific Protein Labeling in Vitro and in Live Mammalian Cells via Rapid Fluorogenic Diels–Alder Reactions. Journal of the American Chemical Society 2012 134 (25), 10317-10320. doi: external page 10.1021/ja302832g
Chayasith Uttamapinant, Jonathan D. Howe, Kathrin Lang, Václav Beránek, Lloyd Davis, Mohan Mahesh, Nicholas P. Barry, and Jason W. Chin (2015): Genetic Code Expansion Enables Live-Cell and Super-Resolution Imaging of Site-Specifically Labeled Cellular Proteins. Journal of the American Chemical Society 2015 137 (14), 4602-4605 doi: external page 10.1021/ja512838z
Nguyen T. A. , Gronauer T. F., Nast-Kolb T. , Sieber S.A , Lang K. (2021): Substrate Profiling of Mitochondrial Caseinolytic Protease P via a Site-Specific Photocrosslinking Approach. Angew. Chem.Int. 2022,61,e202111085. doi: external page 10.1002/anie.202111085
S.V. Mayer, A. Murnauer, M.K. von Wrisberg, M.L. Jokisch and K. Lang (2019): Photo-Induced and Rapid Labeling of Tetrazine-Bearing Proteins via Cyclopropenone-Caged Bicyclononynes. Angew. Chem. Int, 58, 15876-15882 doi: external page 10.1002/anie.201908209
M. Fottner, A-D. Brunner, V. Bittl, D. Horn-Ghetko, A. Jussupow, V.R.I. Kaila, A. Bremm and K. Lang (2019): Site-specific ubiquitylation and SUMOylation using genetic code expansion and sortase. Nat. Chem. Biol. , 15, 276-284 doi: external page 10.1038/s41589-019-0227-4
T.A. Nguyen, M. Cigler, K. Lang. (2018): Expanding the Genetic Code to Study Protein–Protein Interactions Angew. Chem. Int. Ed. 2018, 57, 14350-14361 doi: external page 10.1002/anie.201805869